Subprocess¶
11/17/2022¶
Final Project - Due 11:59pm 12/6/2022¶
Create an assignment, ideally related to your research.
The script resulting from the assignment should be a general purpose tool capable of taking different inputs. You are not required to decompose your assignment into different levels of partial credit (e.g. 70%, 80%, etc.) but you may find it useful to structure it that way for organizational purposes.
You may use any python packages as long as they can be installed with a package manager.
You are required to provide:
- A writeup with sufficient detail for a student to understand and implement the assignment.
- The python code to the solution.
- At least three usage examples
- commandlines with provided inputs and expected outputs
- An in-class presentation describing the assignment (8-10 minutes)
Make sure it is okay to publicly release the data.
Grading Rubric¶
Presentation¶
- Is the problem sufficiently motivated?
- Are relevant and informative visual aids used (avoid massive walls of text!)?
- Is the input and desired output clearly described?
- Is there an outline for how the code will work?
Writeup¶
- Is sufficient, but not excessive, background information provided?
- Is the task clearly described?
- Are the steps required to complete the task presented in a logical manner?
- Are there at least three examples, complete with commandline?
- Are the examples sufficient to demonstrate the task?
Code¶
- Does the code do what it is suppose to?
- Are gross inefficiencies avoided?
- Are appropriate data structures used?
- Are language features appropriately used?
Going Outside the (Python) Box¶
Sometimes you need to integrate with programs that don't have a python interface (or you think it would just be easier to use the command line interface).
Python has a versatile subprocess module for calling and interacting with other programs.
However, first the venerable system command:
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 348 100 348 0 0 4143 0 --:--:-- --:--:-- --:--:-- 4192
0
The return value of the system command is the exit code (not what is printed to screen).
348
subprocess¶
The subprocess module replaces the following modules (so don't use them):
os.system
os.spawn*
os.popen*
popen2.*
commands.*
subprocess.call¶
dump file with spaces ligs.sdf ligs.sdf.1 receptor.pdb receptor.pdb.1 smina
0
Run the command described by ARGS. Wait for command to complete, then return the returncode attribute.
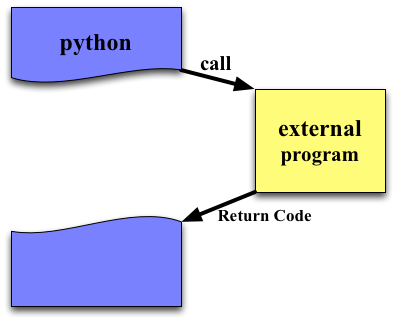
ARGS¶
ARGS specifies the command to call and its arguments. It can either be a string or a list of strings.
0
hello
0
If shell=False (default) and args is a string, it must be only the name of the program (no arguments). If a list is provided, then the first element is the program name and the remaining elements are the arguments.
shell¶
If (and only if) shell = True then the string provided for args is parsed exactly as if you typed it on the commandline. This means you that:
- you must escape special characters (e.g. spaces in file names)
- you can use the wildcard '*' character to expand file names
- you can add IO redirection
If shell=False then list arguments must be use and they are passed literally to the program (e.g., it would get '*' for a file name).
shell is False by default for security reasons. Consider:
filename = input("What file would you like to display?\n")
What file would you like to display?
non_existent; rm -rf / #
subprocess.call("cat " + filename, shell=True) # Uh-oh. This will end badly...By default /bin/sh is used as the shell. You are probably using bash. You can specify what shell to use with the executable argument.
/bin/sh /bin/bash
0
shell Examples¶
ls: cannot access '*': No such file or directory
2
dump file with spaces ligs.sdf ligs.sdf.1 receptor.pdb receptor.pdb.1 smina 0 2 file with spaces 0 file with spaces 0 2
ls: cannot access 'file': No such file or directory ls: cannot access 'with': No such file or directory ls: cannot access 'spaces': No such file or directory ls: cannot access 'file\ with\ spaces': No such file or directory
--------------------------------------------------------------------------- PermissionError Traceback (most recent call last) /tmp/ipykernel_22967/3042384492.py in <module> ----> 1 subprocess.call('ls *') #why is this FileNotFoundError? ~/apps/anaconda3/lib/python3.9/subprocess.py in call(timeout, *popenargs, **kwargs) 347 retcode = call(["ls", "-l"]) 348 """ --> 349 with Popen(*popenargs, **kwargs) as p: 350 try: 351 return p.wait(timeout=timeout) ~/apps/anaconda3/lib/python3.9/subprocess.py in __init__(self, args, bufsize, executable, stdin, stdout, stderr, preexec_fn, close_fds, shell, cwd, env, universal_newlines, startupinfo, creationflags, restore_signals, start_new_session, pass_fds, user, group, extra_groups, encoding, errors, text, umask) 949 encoding=encoding, errors=errors) 950 --> 951 self._execute_child(args, executable, preexec_fn, close_fds, 952 pass_fds, cwd, env, 953 startupinfo, creationflags, shell, ~/apps/anaconda3/lib/python3.9/subprocess.py in _execute_child(self, args, executable, preexec_fn, close_fds, pass_fds, cwd, env, startupinfo, creationflags, shell, p2cread, p2cwrite, c2pread, c2pwrite, errread, errwrite, restore_signals, gid, gids, uid, umask, start_new_session) 1819 if errno_num != 0: 1820 err_msg = os.strerror(errno_num) -> 1821 raise child_exception_type(errno_num, err_msg, err_filename) 1822 raise child_exception_type(err_msg) 1823 PermissionError: [Errno 13] Permission denied: 'ls *'
Input/Output/Error¶
Every process (program) has standard places to write output and read input.
- stdin - standard input is usually from the keyboard
- stdout - standard output is usually buffered
- stderr - standard error is unbuffered (output immediately)
On the commandline, you can changes these places with IO redirection (<,>,|). For example:
grep Congress cnn.html > congress
wc < congress
grep Congress cnn.html | wc
When calling external programs from scripts we'll usually want to provide input to the programs and read their output, so we'll have to change these 'places' as well.
stdin/stdout/stderr¶
stdin, stdout and stderr specify the executed program’s standard input, standard output and standard error file handles, respectively. Valid values are
subprocess.PIPE- this enables communication between your script and the program- an existing file object - e.g. created with
open - None - the program will default to the existing stdin/stdout/stderr
Do no use subprocess.PIPE with subprocess.call
Redirecting to files¶
'dump\n'
2
ls: cannot access 'nonexistantfile': No such file or directory
subprocess.check_call¶
check_call is identical to call, but throws an exception when the called program has a nonzero return value.
ls: cannot access 'missingfile': No such file or directory
--------------------------------------------------------------------------- CalledProcessError Traceback (most recent call last) /tmp/ipykernel_9256/1974956330.py in <module> ----> 1 subprocess.check_call(['ls','missingfile']) ~/apps/anaconda3/lib/python3.9/subprocess.py in check_call(*popenargs, **kwargs) 371 if cmd is None: 372 cmd = popenargs[0] --> 373 raise CalledProcessError(retcode, cmd) 374 return 0 375 CalledProcessError: Command '['ls', 'missingfile']' returned non-zero exit status 2.
subprocess.check_output¶
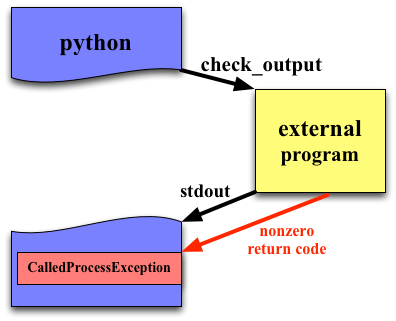
subprocess.check_output¶
b'dump\nfile with spaces\n'
Typically, you are calling a program because you want to parse its output. check_output provides the easiest way to do this. It's return value is what was written to stdout.
Nonzero return values result in a CalledProcessError exception (like check_call).
b'file with spaces\n'
subprocess.check_output¶
Can redirect stderr to STDOUT
b"ls: cannot access 'non_existent_file': No such file or directory\n"
Why exit 0?
b'dump\nfile with spaces\nligs.sdf\nligs.sdf.1\nreceptor.pdb\nreceptor.pdb.1\nsmina\n'
How can we communicate with the program we are launching?

Popen¶
All the previous functions are just convenience wrappers around the Popen object.
dump file with spaces
<Popen: returncode: None args: 'ls'>
Popen has quite a few optional arguments. Shown are just the most common.
cwd sets the working directory for the process (if None defaults to the current working directory of the python script).
env is a dictionary that can be used to define a new set of environment variables.
Popen is a constructor and returns a Popen object.
subprocess.Popen
Popen¶
The python script does not wait for the called process to finish before returning.
We can finally use PIPE.
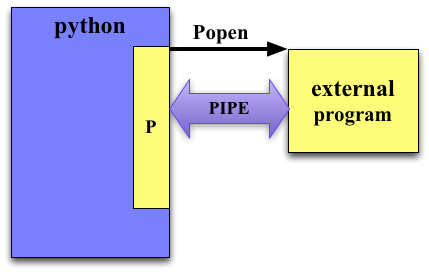
subprocess.PIPE¶
If we set stdin/stdout/stderr to subprocess.PIPE then they are available to read/write to in the resulting Popen object.
_io.BufferedReader
b'dump\n'
subprocess.PIPE¶
Pipes enable communication between your script and the called program.
If stdout/stdin/stderr is set to subprocess.PIPE then that input/output stream of the process is accessible through a file object in the returned object.
b'Hello'
python3 strings are unicode, but most programs need byte strings
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) /tmp/ipykernel_9256/2142223569.py in <module> 1 proc = subprocess.Popen('cat',stdin=subprocess.PIPE,stdout=subprocess.PIPE) ----> 2 proc.stdin.write("Hello") 3 proc.stdin.close() 4 print(proc.stdout.read()) TypeError: a bytes-like object is required, not 'str'
Unicode (aside)¶
Bytes strings (which were the default kinds of string in python2) store each character using a single byte (ASCII, like in the Martian).
Unicode uses 1 to 6 bytes per a character.
This allows supports for other languages and the all important emoji.
💩
Converting bytes to string
'a byte str'
b'a unicode string'
Warning!¶
Managing simultaneous input and output is tricky and can easily lead to deadlocks.
For example, your script may be blocked waiting for output from the process which is blocked waiting for input.

Popen.communicate(input=None)¶
Interact with process: Send data to stdin. Read data from stdout and stderr, until end-of-file is reached. Wait for process to terminate.
input is a string of data to be provided to stdin (which must be set to PIPE).
Likewise, to receive stdout/stderr, they must be set to PIPE.
This will not deadlock.
99% of the time if you have to both provide input and read output of a subprocess, communicate will do what you need.
x 1 a
Interacting with Popen¶
Popen.poll()- check to see if process has terminatedPopen.wait()- wait for process to terminate Do not use PIPEPopen.terminate()- terminate the process (ask nicely)Popen.kill()- kill the process with extreme prejudice
Note that if your are generating a large amount of data, communicate, which buffers all the data in memory, may not be an option (instead just read from Popen.stdout).
If you need to PIPE both stdin and stdout and can't use communicate, be very careful about controlling how data is communicated.
Review¶
- Just want to run a command?
subprocess.call
- Want the output of the command?
subprocess.check_output
- Don't want to wait for command to finish?
subprocess.Popen
- Need to provide data through stdin?
subprocess.Popen,stdin=subprocess.PIPE,communicate
Exercise¶
We want to predict the binding affinity of a small molecule to a protein using the program smina.
--2022-11-16 13:48:38-- https://asinansaglam.github.io/python_bio_2022/files/rec.pdb Resolving asinansaglam.github.io (asinansaglam.github.io)... 185.199.109.153, 185.199.111.153, 185.199.108.153, ... Connecting to asinansaglam.github.io (asinansaglam.github.io)|185.199.109.153|:443... connected. HTTP request sent, awaiting response... 404 Not Found 2022-11-16 13:48:38 ERROR 404: Not Found. --2022-11-16 13:48:38-- https://asinansaglam.github.io/python_bio_2022/files/lig.pdb Resolving asinansaglam.github.io (asinansaglam.github.io)... 185.199.109.153, 185.199.111.153, 185.199.108.153, ... Connecting to asinansaglam.github.io (asinansaglam.github.io)|185.199.109.153|:443... connected. HTTP request sent, awaiting response... 404 Not Found 2022-11-16 13:48:38 ERROR 404: Not Found. --2022-11-16 13:48:38-- https://asinansaglam.github.io/python_bio_2022/files/receptor.pdb Resolving asinansaglam.github.io (asinansaglam.github.io)... 185.199.109.153, 185.199.111.153, 185.199.108.153, ... Connecting to asinansaglam.github.io (asinansaglam.github.io)|185.199.109.153|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 143208 (140K) [application/vnd.palm] Saving to: ‘receptor.pdb.1’ receptor.pdb.1 100%[===================>] 139.85K --.-KB/s in 0.05s 2022-11-16 13:48:39 (2.61 MB/s) - ‘receptor.pdb.1’ saved [143208/143208] --2022-11-16 13:48:39-- https://asinansaglam.github.io/python_bio_2022/files/ligs.sdf Resolving asinansaglam.github.io (asinansaglam.github.io)... 185.199.109.153, 185.199.111.153, 185.199.108.153, ... Connecting to asinansaglam.github.io (asinansaglam.github.io)|185.199.109.153|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 65619 (64K) [application/octet-stream] Saving to: ‘ligs.sdf.1’ ligs.sdf.1 100%[===================>] 64.08K --.-KB/s in 0.02s 2022-11-16 13:48:39 (3.91 MB/s) - ‘ligs.sdf.1’ saved [65619/65619] --2022-11-16 13:48:39-- https://sourceforge.net/projects/smina/files/smina.static/download Resolving sourceforge.net (sourceforge.net)... 104.18.10.128, 104.18.11.128, 2606:4700::6812:a80, ... Connecting to sourceforge.net (sourceforge.net)|104.18.10.128|:443... connected. HTTP request sent, awaiting response... 302 Found Location: https://downloads.sourceforge.net/project/smina/smina.static?ts=gAAAAABjdTCHNXSW3WcCSoVhdsyazoNqg6gJ9sjkOjM57x-YJjgUYUI-QFd3ZRv32ihx1LemPa-FVzFF2M3l1N85LIfwhq7LOQ%3D%3D&use_mirror=cytranet&r= [following] --2022-11-16 13:48:39-- https://downloads.sourceforge.net/project/smina/smina.static?ts=gAAAAABjdTCHNXSW3WcCSoVhdsyazoNqg6gJ9sjkOjM57x-YJjgUYUI-QFd3ZRv32ihx1LemPa-FVzFF2M3l1N85LIfwhq7LOQ%3D%3D&use_mirror=cytranet&r= Resolving downloads.sourceforge.net (downloads.sourceforge.net)... 204.68.111.105 Connecting to downloads.sourceforge.net (downloads.sourceforge.net)|204.68.111.105|:443... connected. HTTP request sent, awaiting response... 302 Found Location: https://cytranet.dl.sourceforge.net/project/smina/smina.static [following] --2022-11-16 13:48:40-- https://cytranet.dl.sourceforge.net/project/smina/smina.static Resolving cytranet.dl.sourceforge.net (cytranet.dl.sourceforge.net)... 162.251.237.20 Connecting to cytranet.dl.sourceforge.net (cytranet.dl.sourceforge.net)|162.251.237.20|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 9853920 (9.4M) [application/octet-stream] Saving to: ‘download’ download 100%[===================>] 9.40M 5.47MB/s in 1.7s 2022-11-16 13:48:42 (5.47 MB/s) - ‘download’ saved [9853920/9853920]
Project¶
- Run the command
smina -r rec.pdb -l lig.pdb --minimizeon these files. Parse the affinity and RMSD and print them on one line. - Run the command
smina -r receptor.pdb -l ligs.sdf --minimize. Parse the affinities and RMSDS.- Plot histograms of both
- Plot a scatter plot
_______ _______ _________ _ _______
( ____ \( )\__ __/( ( /|( ___ )
| ( \/| () () | ) ( | \ ( || ( ) |
| (_____ | || || | | | | \ | || (___) |
(_____ )| |(_)| | | | | (\ \) || ___ |
) || | | | | | | | \ || ( ) |
/\____) || ) ( |___) (___| ) \ || ) ( |
\_______)|/ \|\_______/|/ )_)|/ \|
smina is based off AutoDock Vina. Please cite appropriately.
Weights Terms
-0.035579 gauss(o=0,_w=0.5,_c=8)
-0.005156 gauss(o=3,_w=2,_c=8)
0.840245 repulsion(o=0,_c=8)
-0.035069 hydrophobic(g=0.5,_b=1.5,_c=8)
-0.587439 non_dir_h_bond(g=-0.7,_b=0,_c=8)
1.923 num_tors_div
Affinity: -6.13684 -0.44100 (kcal/mol)
RMSD: 0.04666
Refine time 0.01145
Affinity: -5.86570 -0.18850 (kcal/mol)
RMSD: 0.16984
Refine time 0.00334
Affinity: -6.05768 -1.30419 (kcal/mol)
RMSD: 0.07613
Refine time 0.00347
Affinity: -6.59074 -0.53131 (kcal/mol)
RMSD: 0.22924
Refine time 0.00641
Affinity: -6.50168 0.08280 (kcal/mol)
RMSD: 0.04795
Refine time 0.00282
Affinity: -5.88335 -0.73565 (kcal/mol)
RMSD: 0.07531
Refine time 0.00204
Affinity: -6.94803 -0.27693 (kcal/mol)
RMSD: 0.08882
Refine time 0.00816
Affinity: -6.11432 -0.32757 (kcal/mol)
RMSD: 0.06710
Refine time 0.00543
Affinity: -5.85392 -0.32171 (kcal/mol)
RMSD: 0.98681
Refine time 0.00488
Affinity: -6.80549 -0.59248 (kcal/mol)
RMSD: 0.17517
Refine time 0.00431
Affinity: -6.73040 -0.57962 (kcal/mol)
RMSD: 0.03489
Refine time 0.00198
Affinity: -5.69268 -0.47264 (kcal/mol)
RMSD: 0.14554
Refine time 0.00318
Affinity: -5.08187 -2.76936 (kcal/mol)
RMSD: 0.10448
Refine time 0.00329
Affinity: -6.44079 -0.76932 (kcal/mol)
RMSD: 0.04434
Refine time 0.00256
Affinity: -6.45828 -0.45417 (kcal/mol)
RMSD: 0.09374
Refine time 0.00619
Affinity: -6.65080 -0.63172 (kcal/mol)
RMSD: 0.08727
Refine time 0.00643
Affinity: -7.12596 -0.32647 (kcal/mol)
RMSD: 0.14559
Refine time 0.00377
Affinity: -6.77129 -0.58484 (kcal/mol)
RMSD: 0.21863
Refine time 0.00683
Affinity: -7.54122 -1.03283 (kcal/mol)
RMSD: 0.06561
Refine time 0.00443
Affinity: -5.62031 -0.34329 (kcal/mol)
RMSD: 0.22742
Refine time 0.00298
Affinity: -6.35736 -0.69922 (kcal/mol)
RMSD: 0.12231
Refine time 0.00419
Affinity: -5.79781 -0.80878 (kcal/mol)
RMSD: 0.14716
Refine time 0.00204
Affinity: -5.88094 -0.42970 (kcal/mol)
RMSD: 0.11252
Refine time 0.00260
Affinity: -7.09409 0.36596 (kcal/mol)
RMSD: 0.41997
Refine time 0.00359
Affinity: -6.13325 -0.22617 (kcal/mol)
RMSD: 0.08001
Refine time 0.00307
Affinity: -7.47566 -1.20172 (kcal/mol)
RMSD: 0.35906
Refine time 0.00858
Affinity: -6.47657 -0.41204 (kcal/mol)
RMSD: 0.04145
Refine time 0.00277
Affinity: -6.58339 -0.62376 (kcal/mol)
RMSD: 0.22803
Refine time 0.00525
Affinity: -6.69025 -0.11960 (kcal/mol)
RMSD: 0.15410
Refine time 0.00368
Affinity: -5.73675 -0.69679 (kcal/mol)
RMSD: 0.06322
Refine time 0.00217
Loop time 0.16327